This function implements the landmark Aalen-Johansen method of Putter & Spitoni (2016) for non-parametric estimation of transition probabilities in non-Markov models.
Usage
LMAJ(msdata, s, from, method = c("aalen", "greenwood"))Arguments
- msdata
An
"msdata"object, as for instance prepared bylink{msprep}- s
The prediction time point s from which transition probabilities are to be obtained
- from
Either a single state or a set of states in the state space 1,...,S
- method
The method for calculating variances, as in
probtrans
Value
A data frame containing estimates and associated standard errors of
the transition probabilities P(X(t)=k | X(s) in from) with s
and from the arguments of the function.
References
H. Putter and C. Spitoni (2016). Estimators of transition probabilities in non-Markov multi-state models. Submitted.
Examples
data(prothr)
tmat <- attr(prothr, "trans")
pr0 <- subset(prothr, treat=="Placebo")
attr(pr0, "trans") <- tmat
pr1 <- subset(prothr, treat=="Prednisone")
attr(pr1, "trans") <- tmat
c0 <- coxph(Surv(Tstart, Tstop, status) ~ strata(trans), data=pr0)
#> Warning: Stop time must be > start time, NA created
c1 <- coxph(Surv(Tstart, Tstop, status) ~ strata(trans), data=pr1)
#> Warning: Stop time must be > start time, NA created
msf0 <- msfit(c0, trans=tmat)
#> Warning: Stop time must be > start time, NA created
#> Warning: Stop time must be > start time, NA created
msf1 <- msfit(c1, trans=tmat)
#> Warning: Stop time must be > start time, NA created
#> Warning: Stop time must be > start time, NA created
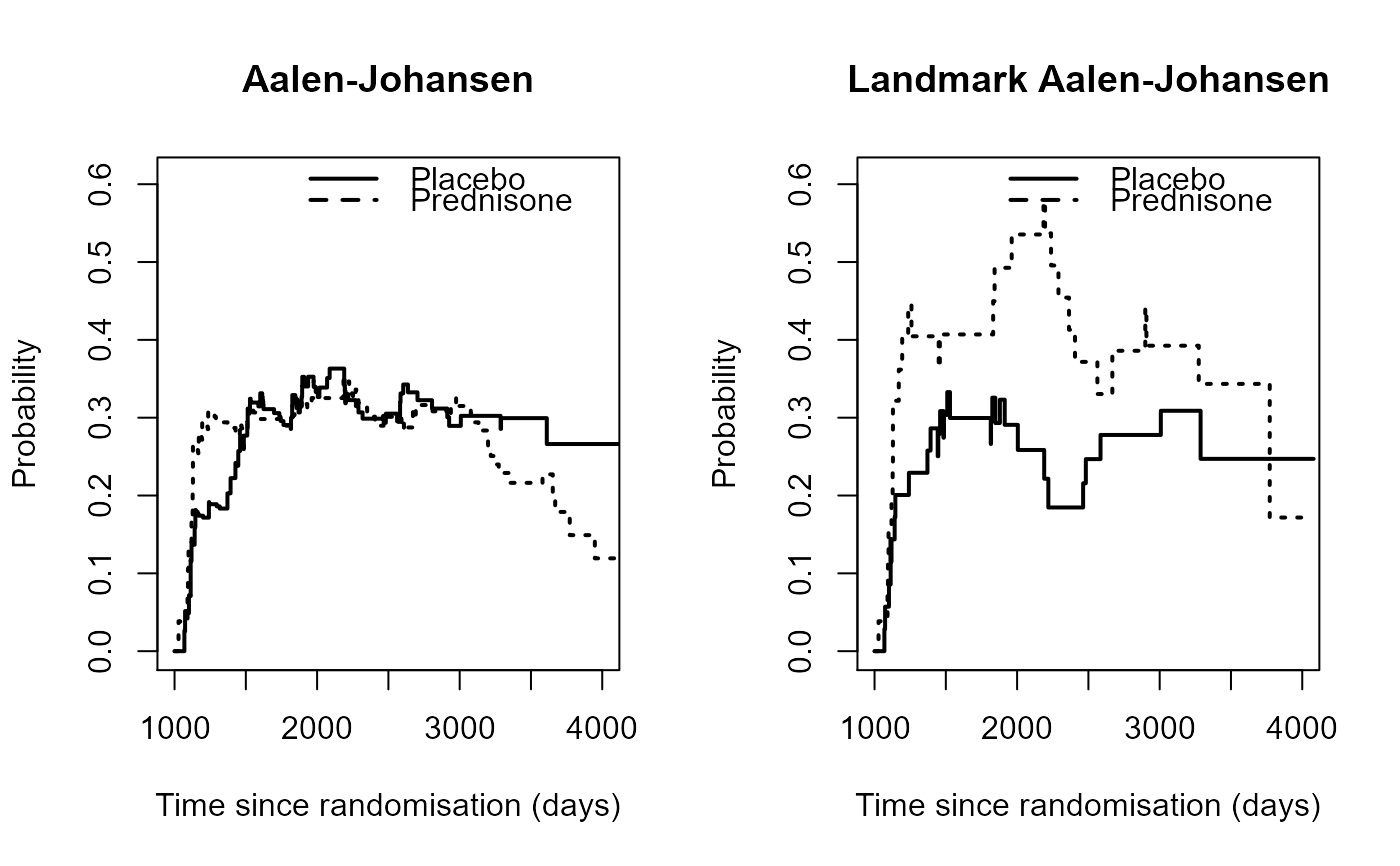
# Comparison as in Figure 2 of Titman (2015)
# Aalen-Johansen
pt0 <- probtrans(msf0, predt=1000)[[2]]
pt1 <- probtrans(msf1, predt=1000)[[2]]
par(mfrow=c(1,2))
plot(pt0$time, pt0$pstate1, type="s", lwd=2, xlim=c(1000,4000), ylim=c(0,0.61),
xlab="Time since randomisation (days)", ylab="Probability")
lines(pt1$time, pt1$pstate1, type="s", lwd=2, lty=3)
legend("topright", c("Placebo", "Prednisone"), lwd=2, lty=1:2, bty="n")
title(main="Aalen-Johansen")
# Landmark Aalen-Johansen
LMpt0 <- LMAJ(msdata=pr0, s=1000, from=2)
#> Warning: Stop time must be > start time, NA created
#> Warning: Stop time must be > start time, NA created
#> Warning: Stop time must be > start time, NA created
#> Warning: no non-missing arguments to max; returning -Inf
LMpt1 <- LMAJ(msdata=pr1, s=1000, from=2)
#> Warning: Stop time must be > start time, NA created
#> Warning: Stop time must be > start time, NA created
#> Warning: Stop time must be > start time, NA created
#> Warning: no non-missing arguments to max; returning -Inf
plot(LMpt0$time, LMpt0$pstate1, type="s", lwd=2, xlim=c(1000,4000), ylim=c(0,0.61),
xlab="Time since randomisation (days)", ylab="Probability")
lines(LMpt1$time, LMpt1$pstate1, type="s", lwd=2, lty=3)
legend("topright", c("Placebo", "Prednisone"), lwd=2, lty=1:2, bty="n")
title(main="Landmark Aalen-Johansen")