Plot method for an object of class "msfit". It plots the estimated
cumulative transition intensities in the multi-state model.
Arguments
- x
Object of class
"msfit", containing estimated cumulative transition intensities for all transitions in a multi-state model- type
One of
"single"(default) or"separate"; in case of"single", all estimated cumulative hazards are drawn in a single plot, in case of"separate", separate plots are shown for the estimated transition intensities- cols
A vector specifying colors for the different transitions; default is 1:K (K no of transitions), when type=
"single", and 1 (black), when type="separate"- xlab
A title for the x-axis; default is
"Time"- ylab
A title for the y-axis; default is
"Cumulative hazard"- ylim
The y limits of the plot(s); if ylim is specified for type="separate", then all plots use the same ylim for y limits
- lwd
The line width, see
par; default is 1- lty
The line type, see
par; default is 1- legend
Character vector of length equal to the number of transitions, to be used in a legend; if missing, these will be taken from the row- and column-names of the transition matrix contained in
x$trans. Also used as titles of plots for type="separate"- legend.pos
The position of the legend, see
legend; default is"topleft"- bty
The box type of the legend, see
legend- use.ggplot
Default FALSE, set TRUE for ggplot version of plot
- xlim
Limits of x axis, relevant if use_ggplot = T
- scale_type
"fixed", "free", "free_x" or "free_y", see scales argument of facet_wrap(). Only relevant for use_ggplot = T.
- conf.int
Confidence level (%) from 0-1 for the cumulative hazard, default is 0.95. Only relevant for use_ggplot = T
- conf.type
Type of confidence interval - either "log" or "plain" . See function details of
plot.probtransfor details- ...
Further arguments to plot
Examples
# transition matrix for illness-death model
tmat <- trans.illdeath()
# data in wide format, for transition 1 this is dataset E1 of
# Therneau & Grambsch (2000)
tg <- data.frame(illt=c(1,1,6,6,8,9),ills=c(1,0,1,1,0,1),
dt=c(5,1,9,7,8,12),ds=c(1,1,1,1,1,1),
x1=c(1,1,1,0,0,0),x2=c(6:1))
# data in long format using msprep
tglong <- msprep(time=c(NA,"illt","dt"),status=c(NA,"ills","ds"),
data=tg,keep=c("x1","x2"),trans=tmat)
# events
events(tglong)
#> $Frequencies
#> to
#> from healthy illness death no event total entering
#> healthy 0 4 2 0 6
#> illness 0 0 4 0 4
#> death 0 0 0 6 6
#>
#> $Proportions
#> to
#> from healthy illness death no event
#> healthy 0.0000000 0.6666667 0.3333333 0.0000000
#> illness 0.0000000 0.0000000 1.0000000 0.0000000
#> death 0.0000000 0.0000000 0.0000000 1.0000000
#>
table(tglong$status,tglong$to,tglong$from)
#> , , = 1
#>
#>
#> 2 3
#> 0 2 4
#> 1 4 2
#>
#> , , = 2
#>
#>
#> 2 3
#> 0 0 0
#> 1 0 4
#>
# expanded covariates
tglong <- expand.covs(tglong,c("x1","x2"))
# Cox model with different covariate
cx <- coxph(Surv(Tstart,Tstop,status)~x1.1+x2.2+strata(trans),
data=tglong,method="breslow")
summary(cx)
#> Call:
#> coxph(formula = Surv(Tstart, Tstop, status) ~ x1.1 + x2.2 + strata(trans),
#> data = tglong, method = "breslow")
#>
#> n= 16, number of events= 10
#>
#> coef exp(coef) se(coef) z Pr(>|z|)
#> x1.1 1.4753 4.3723 1.2557 1.175 0.240
#> x2.2 0.8571 2.3563 0.8848 0.969 0.333
#>
#> exp(coef) exp(-coef) lower .95 upper .95
#> x1.1 4.372 0.2287 0.3731 51.24
#> x2.2 2.356 0.4244 0.4160 13.35
#>
#> Concordance= 0.781 (se = 0.077 )
#> Likelihood ratio test= 2.93 on 2 df, p=0.2
#> Wald test = 2.32 on 2 df, p=0.3
#> Score (logrank) test = 2.86 on 2 df, p=0.2
#>
# new data, to check whether results are the same for transition 1 as
# those in appendix E.1 of Therneau & Grambsch (2000)
newdata <- data.frame(trans=1:3,x1.1=c(0,0,0),x2.2=c(0,1,0),strata=1:3)
msf <- msfit(cx,newdata,trans=tmat)
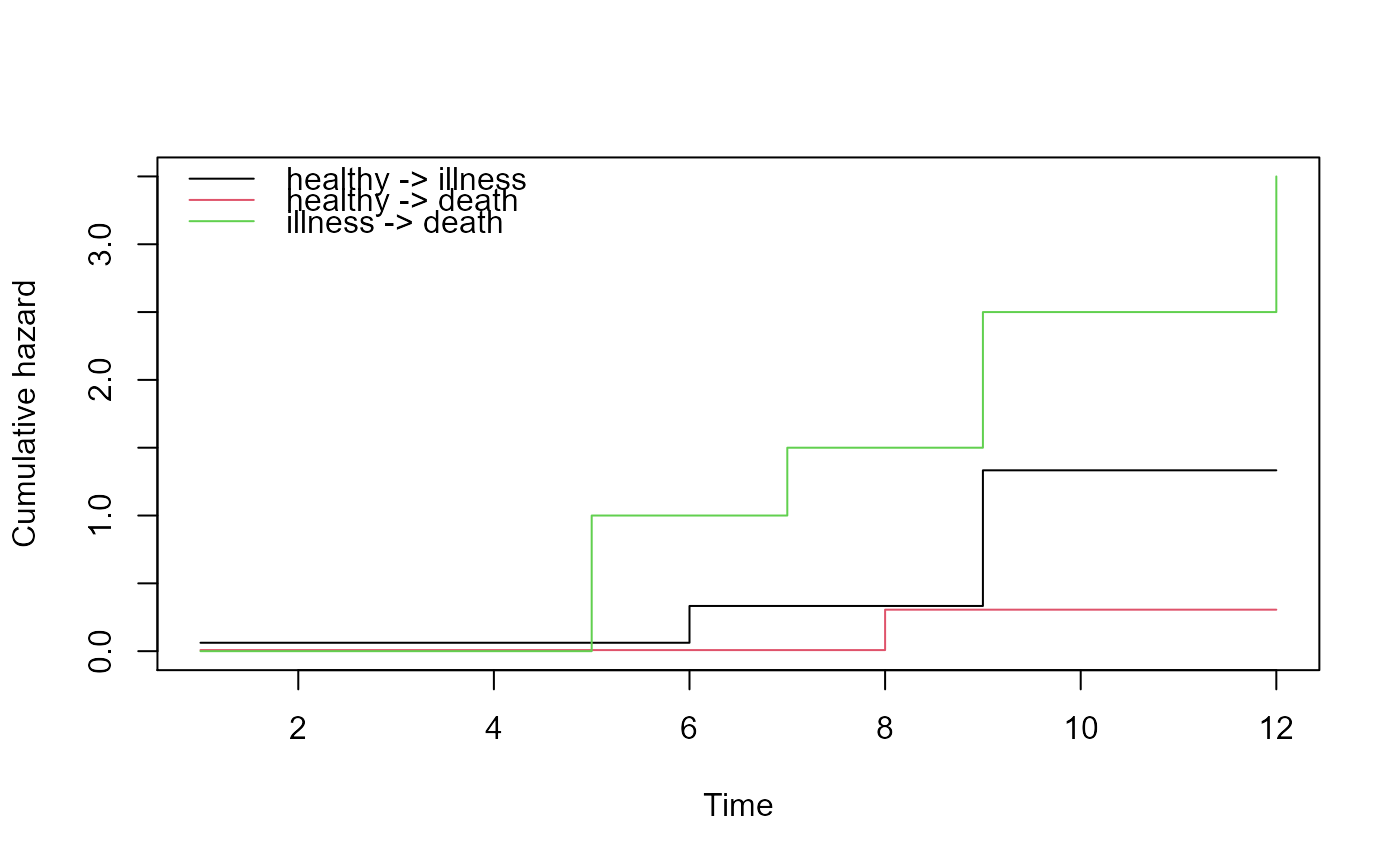
# standard plot
plot(msf)
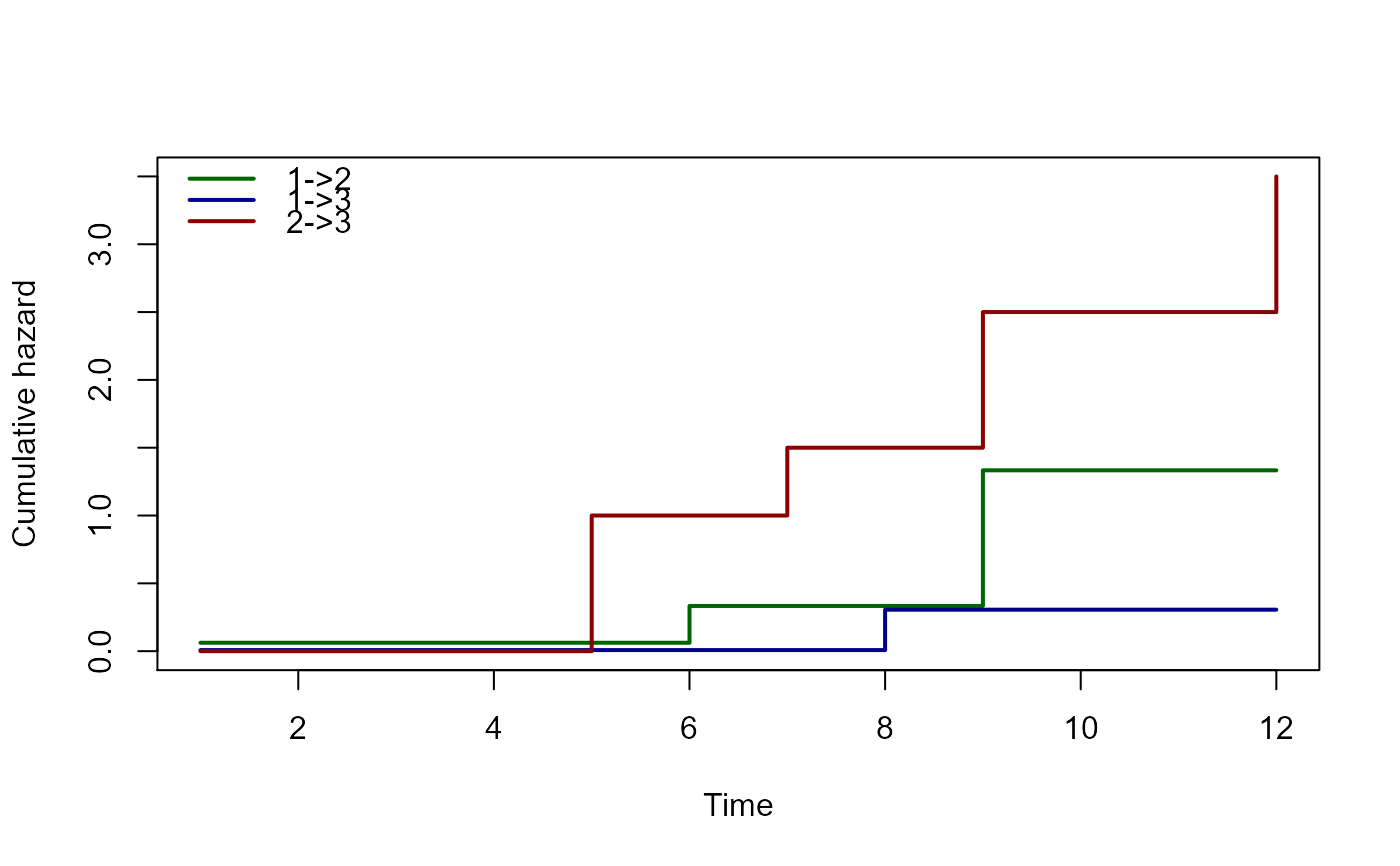
 # specifying line width, color, and legend
plot(msf,lwd=2,col=c("darkgreen","darkblue","darkred"),legend=c("1->2","1->3","2->3"))
# specifying line width, color, and legend
plot(msf,lwd=2,col=c("darkgreen","darkblue","darkred"),legend=c("1->2","1->3","2->3"))
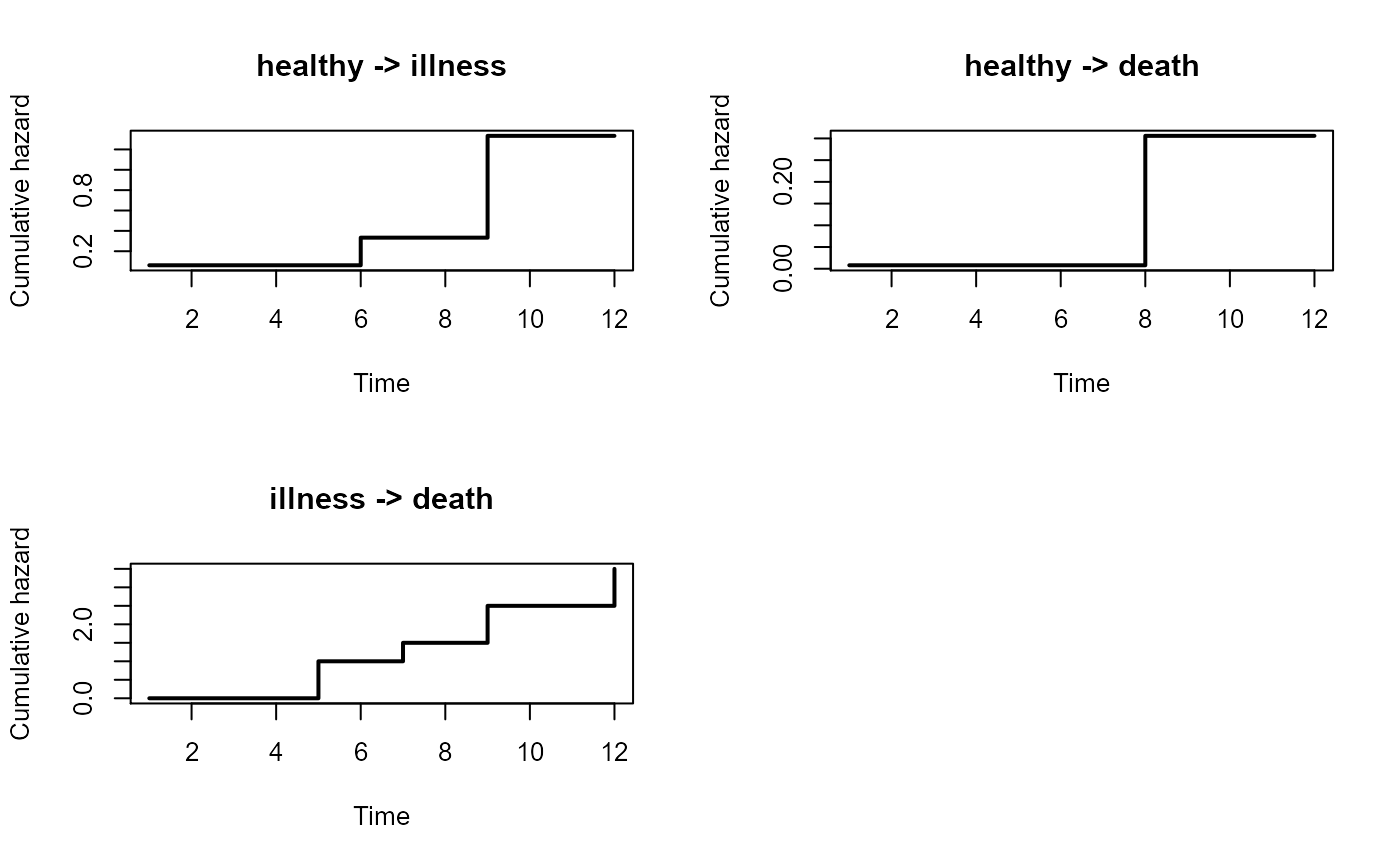
 # separate plots
par(mfrow=c(2,2))
plot(msf,type="separate",lwd=2)
par(mfrow=c(1,1))
# separate plots
par(mfrow=c(2,2))
plot(msf,type="separate",lwd=2)
par(mfrow=c(1,1))
 # ggplot version - see vignette for details
library(ggplot2)
plot(msf, use.ggplot = TRUE)
# ggplot version - see vignette for details
library(ggplot2)
plot(msf, use.ggplot = TRUE)
